Stratigraphic Paleobiology for Phenotypic Evolution
Source:vignettes/phenotypic_evolution.Rmd
phenotypic_evolution.RmdIntroduction
Here we describe the modeling pipeline for the stratigraphic
paleobiology of phenotypic evolution. For details how this can be
combined with the paleoTS package (Hunt 2006), see
vignette("paleoTS_functionality")First, let’s load all required packages:
Modes of evolution in the time domain
Implemented modes of evolution
The StratPal package provides three functions for
simulating different modes of evolution in the time domain:
stasissimulates evolutionary stasis as independent, normally distributed random variables with meanmeanand standard deviationsd(Hunt 2006).random_walksimulates trait evolution following a (potentially biased) random walk with variabilitysigma, directionalitymu, and initial trait valuey0(e.g. Bookstein 1987). Settingmuto a negative (positive) value will make the value of the random walk decrease (increase) over time,sigmadetermines the effect of randomness on the trait values.ornstein_uhlenbeckcorresponds to convergence to a phenotypic optimum, wheremuis the optimal value (long term mean),thetadetermines how fastmuis approached,sigmathe noise level, andy0the initial trait value (Lande 1976).
Use vignette("advanced_functionality") for guidelines to
implement your own modes of evolution.
Visualization
You can visualize the different modes of evolution using the following pipeline:
seq(0, 1, by = 0.01) |> # times of simulation in Myr. Simulate over 1 Myr years with 10 kyr resolution
random_walk(sigma = 1, mu = 3) |> # simulate random walk with increasing trait values
plot(type = "l", # plot results
xlab = "Time [Myr]",
ylab = "Trait value")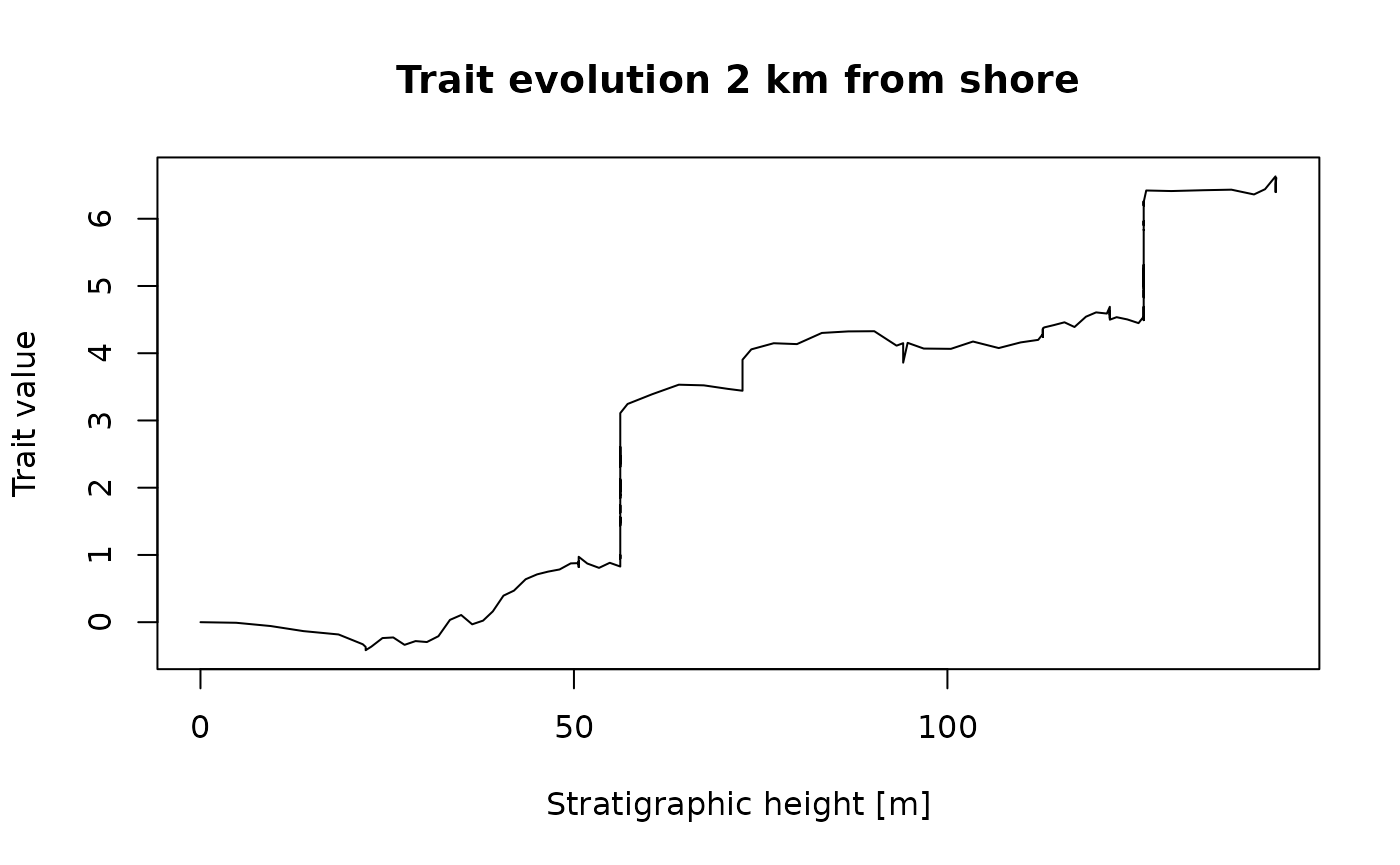
Task: Modify the pipeline to get an intuition for the different modes of evolution. What are the effects of their parameters, and what is their biological meaning?
Stratigraphic domain
We are interested in how phenotypic evolution is preserved in the stratigraphic record, i.e. how trait evolution within a lineage is preserved at a specific location in the stratigraphic record. Here we develop the modeling pipelines to answer this question.
Age-depth models
As an example, we compare the preservation of trait evolution 2 km
and 12 km offshore in the carbonate platform in scenario A from Hohmann et al. (2024). See
?scenarioA and vignette("StratPal") for
details on scenario A.
First define the age-depth models:
adm_2km = tp_to_adm(t = scenarioA$t_myr, # 2 km from shore
h = scenarioA$h_m[,"2km"],
T_unit = "Myr",
L_unit = "m")
adm_12km = tp_to_adm(t = scenarioA$t_myr, # 12 km from shore
h = scenarioA$h_m[,"12km"],
T_unit = "Myr",
L_unit = "m")
plot(adm_2km, # plot age-depth model 2 km from shore
lwd_acc = 2, # plot thicker lines for intervals with sediment accumulation (lwd = line width)
lty_destr = 0) # don't plot destructive intervals/gaps (lty = line type)
T_axis_lab() # add time axis label
L_axis_lab() # add length axis label
title("Age-depth model 2 km from shore")
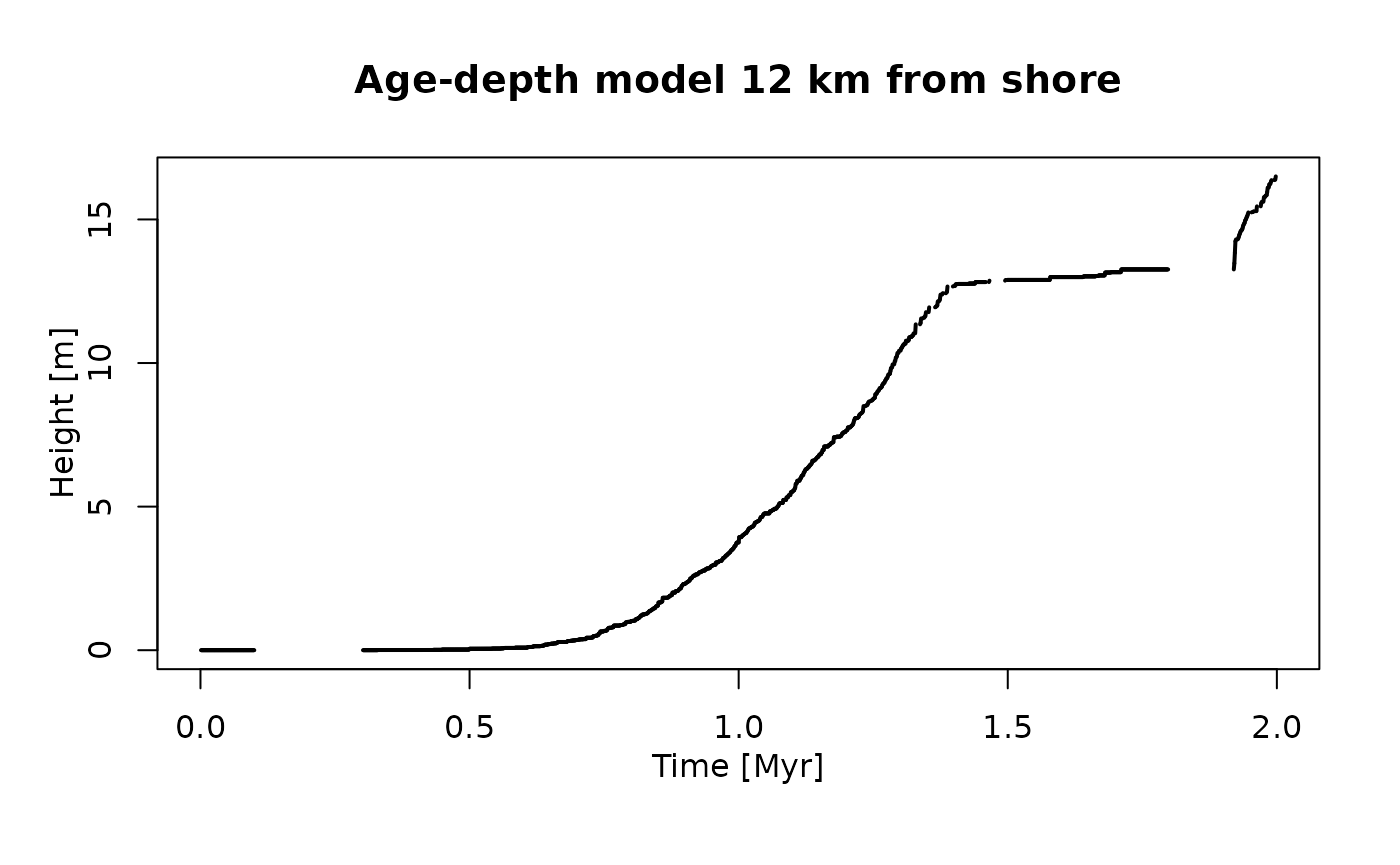
plot(adm_12km, # plot age-depth model 12 km from shore
lwd_acc = 2,
lty_destr = 0)
T_axis_lab()
L_axis_lab()
title("Age-depth model 12 km from shore")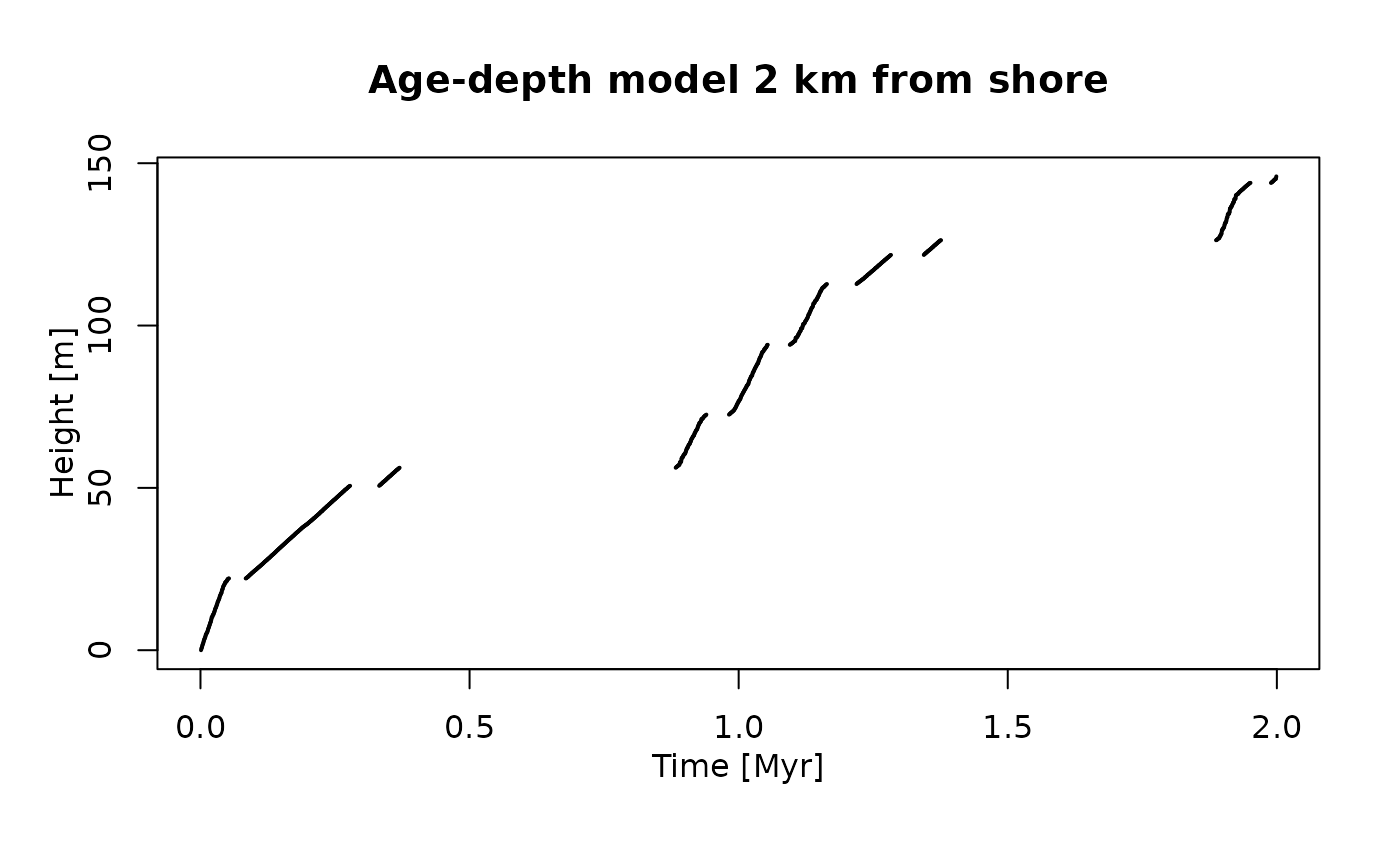
Stratigraphic expression of phenotypic evolution
We use the defined age-depth models to transform the simulated trait
values from the time domain into the depth domain via
time_to_strat:
seq(from = min_time(adm_2km), to = max_time(adm_2km), by = 0.01) |> # sample every 10 kyr over the interval covered by the adm
random_walk(sigma = 1, mu = 3) |> # simulate random walk
time_to_strat(adm_2km, destructive = FALSE) |> # transform data from time to strat domain
plot(type = "l", # plot
orientation = "lr",
xlab = paste0("Stratigraphic height [", get_L_unit(adm_2km), "]"),
ylab = "Trait value",
main = "Trait evolution 2 km from shore")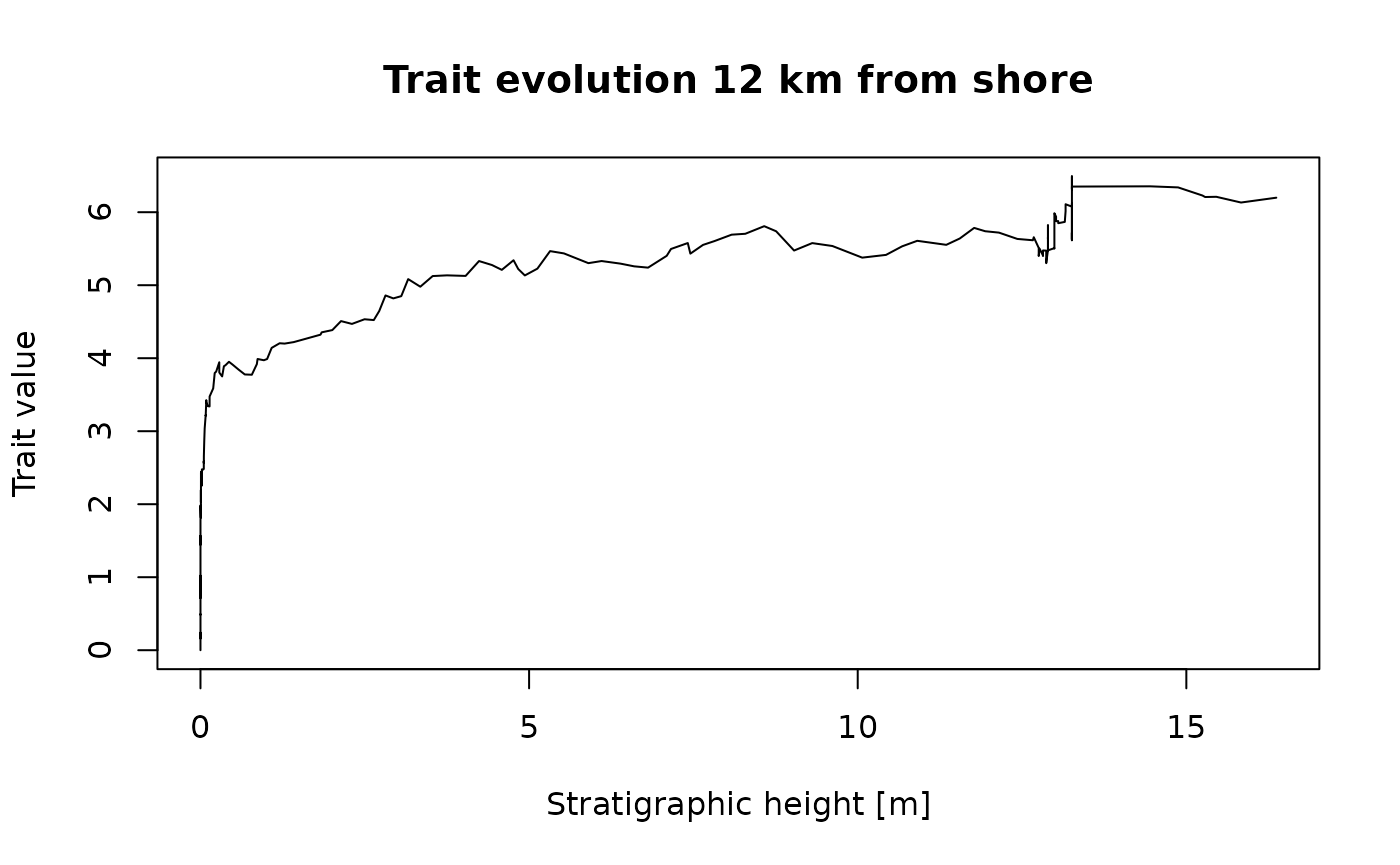
This is what a biased random walk in the time domain would look like if it was observed 2 km offshore in the simulated carbonate platform. The large jumps in traits correspond to long hiatuses caused by prolonged drops in relative sea level (Hohmann et al. 2024). Compare this figure to the random walk in the time domain (without stratigraphic distortions).
12 km offshore, the preservation is very different:
seq(from = min_time(adm_12km), to = max_time(adm_12km), by = 0.01) |> # sample every 10 kyr over the interval covered by the adm
random_walk(sigma = 1, mu = 3) |> # simulate random walk
time_to_strat(adm_12km, destructive = FALSE) |> # transform data from time to strat domain
plot(type = "l", # plot results
orientation = "lr",
xlab = paste0("Stratigraphic height [", get_L_unit(adm_12km), "]"),
ylab = "Trait value",
main = "Trait evolution 12 km from shore")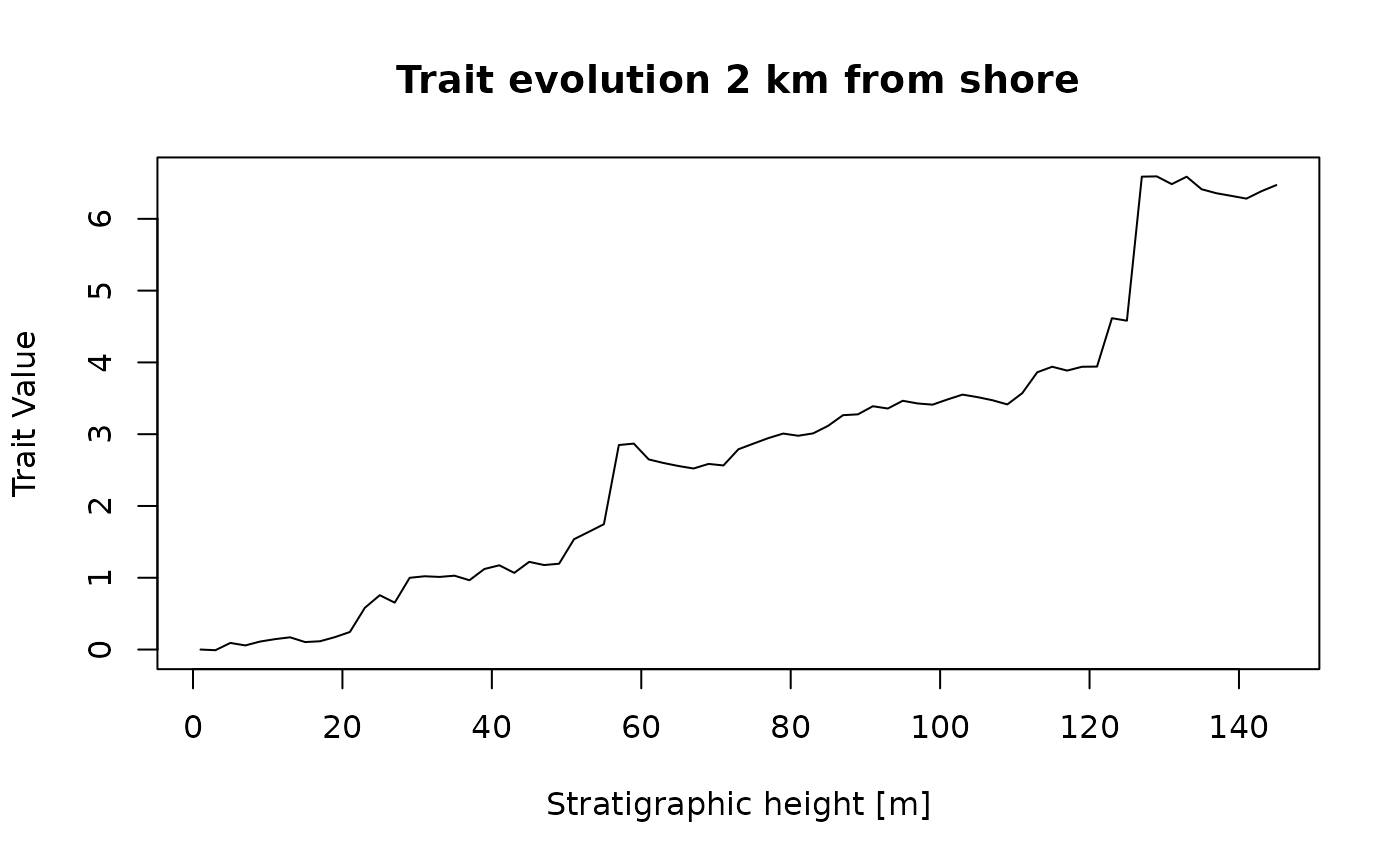
You can also see two jumps, but the first is at a different location compared to the section 2 km offshore. Looking at the age-depth models, we can see why this is: 2 km offshore, the jumps are caused by prolonged hiatuses that are associated with the massive drops in sea level around 0.5 Myr and 1.5 Myr. 12 km offshore, the age-depth model shows fewer gaps, but prolonged intervals with low sedimentation rates at the beginning of the simulation and at around 1.5 Myr. This leads to intervals where the rate phenotypic evolution appears to be accelerated because of (sedimentological) condensation (Hohmann 2021). You can see that not all artefactual jumps in traits observable in the stratigraphic record are caused by gaps.
Task: How does this effect vary between different modes of evolution (with different parameter choices), and at different locations in the platform (e.g., along an onshore-offshore gradient)? Can you make any general statements about where you see the strongest effects?
Prescribing a sampling strategy
The above plots already give us a good idea of how stratigraphic effects can change our interpretation of trait evolution.
However, there is a small imperfection. Because we simulate the lineage every 10 kyr, it is sampled irregularly in the stratigraphic domain: If sedimentation rates are high, samples can be multiple meters apart, but if sedimentation rates are low, they are only a few centimeters apart.
To fix this, we want to prescribe where in the stratigraphic domain samples are taken. This allows us to examine the effect that different sampling strategies have on our perception of trait evolution. We do this in the following steps:
- Determine sampling locations in the stratigraphic domain.
- Calculate what times correspond to said sampling locations via
strat_to_time. - Simulate a lineage at said times.
- Transform the simulated trait values back into the stratigraphic
domain using
time_to_strat. - Plot the result.
Let’s assume we take a sample every 2 meters. Then our five steps above result in the following modeling pipeline:
dist_between_samples_m = 2
sampling_loc_m = seq(from = 0.5 * dist_between_samples_m,
to = max_height(adm_2km),
by = dist_between_samples_m)
sampling_loc_m |> # sampling locations
strat_to_time(adm_2km) |> # determine times where lineage is sampled
random_walk(sigma = 1, mu = 3) |> # simulate trait values at these times
time_to_strat(adm_2km, destructive = FALSE) |> # transform trait values to stratigraphic domain
plot(orientation = "lr", # plot stratigraphic data
type = "l",
ylab = "Trait Value",
xlab = paste0("Stratigraphic height [", get_L_unit(adm_2km), "]"),
main = "Trait evolution 2 km from shore")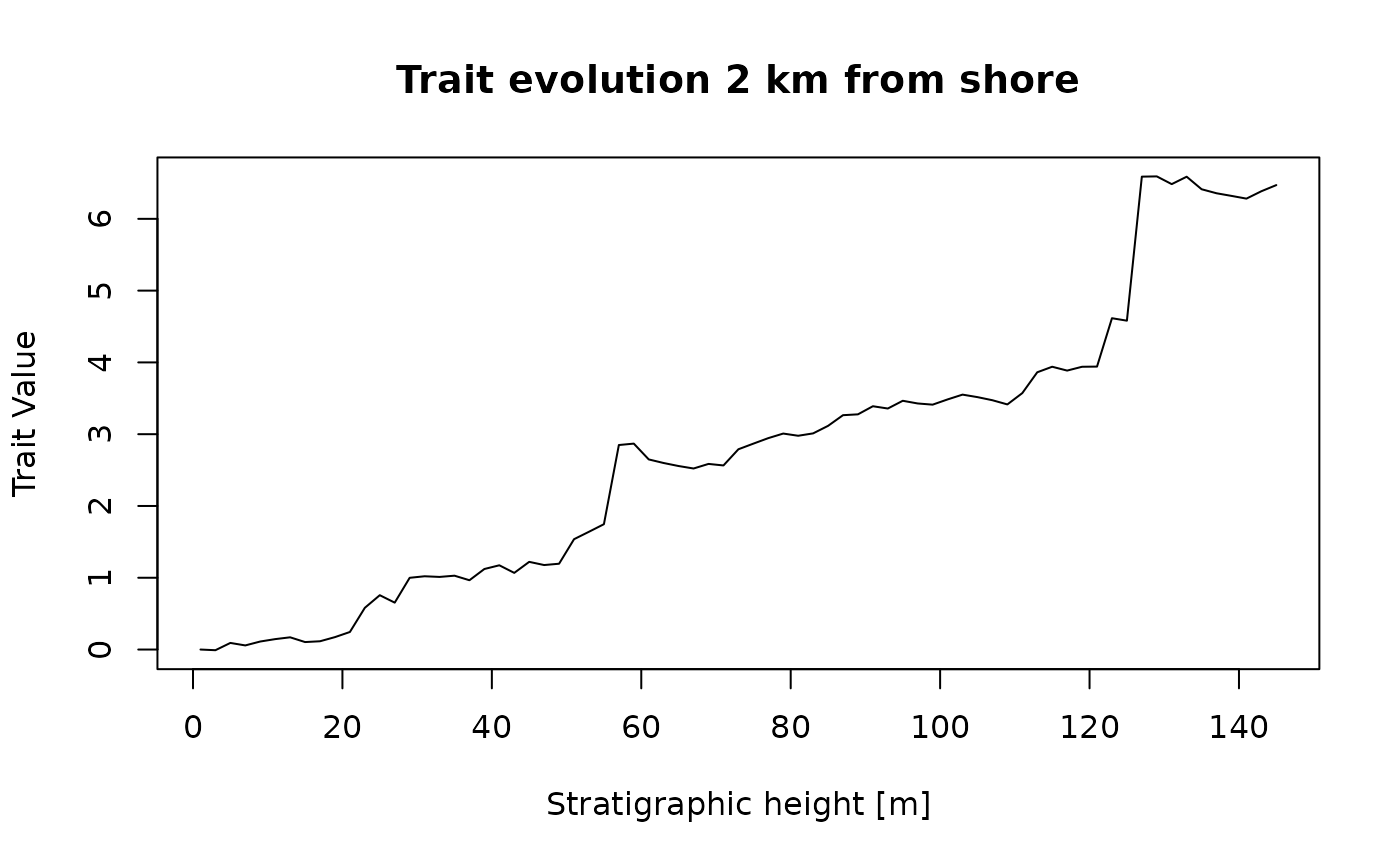
There we have it! This is what the lineage would look like if it was sampled every 2 meters in a sections 2 km from shore.
Task: How does prescribing a sampling strategy change the interpretations of your last task? Can you draw any conclusions about which sampling strategy is best suited for a given environment or mode of evolution?
Further reading
For details on how the paleoTS and evoTS
packages (Hunt 2006, Voje 2023) can be
integrated with the StratPal package, see
vignette("paleoTS_functionality")Run
vignette("event_data")for details on how to model stratigraphic paleobiology of event data such as individual fossils and first/last occurrences of taxa, or explore the vignette online under mindthegap-erc.github.io/StratPal/articles/event_data.
See also
vignette("advanced_functionality")for details on how to expand on the modeling pipelines described here, or explore the vignette online under mindthegap-erc.github.io/StratPal/articles/advanced_functionality.
References
Bookstein, Fred L. 1987. “Random walk and the existence of evolutionary rates.” Paleobiology. https://doi.org/10.1017/S0094837300009039.
Hohmann, Niklas. 2021. “Incorporating information on varying sedimentation rates into paleontological analyses.” PALAIOS. https://doi.org/10.2110/palo.2020.038.
Hohmann, Niklas; Koelewijn, Joël R.; Burgess, Peter; Jarochowska, Emilia. 2024. “Identification of the mode of evolution in incomplete carbonate successions”. BMC Ecology and Evolution 24, 113. https://doi.org/10.1186/s12862-024-02287-2.
Hunt, Gene. 2006. “Fitting and Comparing Models of Phyletic Evolution: Random Walks and Beyond.” Paleobiology. https://doi.org/10.1666/05070.1.
Lande, Russell. 1976. “Natural selection and random genetic drift in phenotypic evolution.” Evolution. https://doi.org/10.1111/j.1558-5646.1976.tb00911.x.
Voje, Kjetil L. 2023. “Fitting and Evaluating Univariate and Multivariate Models of Within-Lineage Evolution.” Paleobiology. https://doi.org/10.1017/pab.2023.10.